The first SpatialData hackathon and workshop was organized by the scverse community on 12-14 November 2024 in Basel, Switzerland. 20 attendees had gathered to work on improving online materials, tutorials, and scalability of existing spatial omics and scverse frameworks where interoperability across software platforms were prioritized.
Interoperability for Spatial Omics
One of the key objectives of the workshop was to gather communities from R and Python, to harmonize spatial data formats towards R/Python interoperability and cross-framework integration.
A group of R/Bioconductor developers led by Helena L. Crowell have teamed up to devise R bindings for the SpatialData framework which will enable users to operate SpatialData objects across R and Python environments simultaneously.
R Interoperability Team (clockwise): Artür Manukyan, Dario Righelli, Vincent Carey, Kevin Yamauchi, Louise Deconinck, Estella Yixing Dong, Helena L. Crowell
The event brought together scientists with expertise in (i) R and Bioconductor frameworks for spatial omics, (ii) R/Python interoperability for single cell frameworks (anndataR) and (iii) Zarr for the storage of chunked, compressed, N-dimensional arrays (Rarr, pizzarr).
Further discussions were made on joint scverse/Bioconductor projects and how to fully utilize R-native frameworks for out of memory data management through file formats such as .zarr (via DelayedArray) and .parquet (via arrow) to scale up spatial omics analysis for the Bioconductor community.
Summary of Hackathon
The Hackathon event in Basel focused on four tracks which leveraged multiple programming languages, including Python, R, and JavaScript:
R interoperability: Enhancing the integration and compatibility of R and Python with the SpatialData Python framework by using the language-agnostic SpatialData Zarr file format.
Visualization interoperability: Improving the seamless integration of visualization tools across different systems and programming languages via a tool-agnostic view configuration.
Scalability and benchmarking: Identifying, benchmarking, and addressing computational bottlenecks within the SpatialData framework.
Ergonomics and user-friendliness: Enhancing the usability and accessibility of the SpatialData framework for both first-time users and third-party developers.
You can find more information on outcomes of each of these tracks in the BioHackArxiv preprint.
For track 1 specifically, R/Bioconductor packages SpatialData (representation), SpatialData.plot (visualization), SpatialData.data (datasets) are under development, and a BiocBook consolidation vignettes and scientific workflows is in planning.
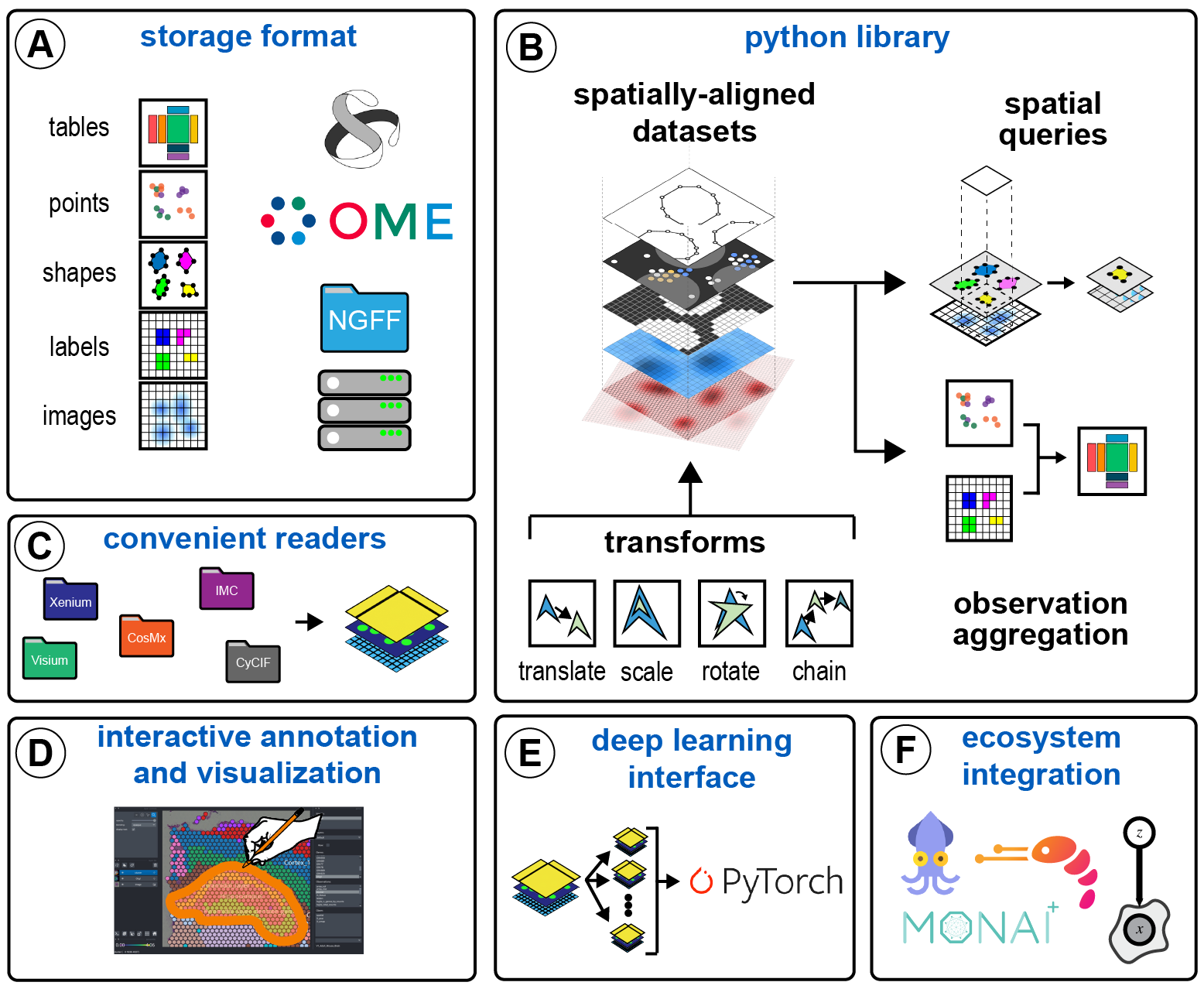
SpatialData Framework
SpatialData is a data framework that comprises a FAIR storage format and a collection of libraries originally written in python for performant access, alignment, and processing of spatial omics datasets. SpatialData is currently maintained by the scverse consortium.
Website: https://spatialdata.scverse.org/
SpatialData core team: Luca Marconato, Giovanni Palla, Kevin Yamauchi, Wouter-Michiel Vierdag, Tim Treis, Isaac Virshup, Josh Moore.
scverse
scverse is a consortium of foundational tools (mostly in Python) for omics data in life sciences. It has been founded to ensure the long-term maintenance of some known core tools such as anndata, scanpy, squidpy and spatialdata.
Website: https://scverse.org/
Join Us!
Join the #spatialdata-devel channel in Bioconductor Slack to take part in discussions, future work, and hackathons.
© 2025 Bioconductor. Content is published under Creative Commons CC-BY-4.0 License for the text and BSD 3-Clause License for any code. | R-Bloggers