Introduction
GPU acceleration enables faster and more scalable analysis workflows, especially for tasks like deep learning, image processing, and large-scale genomics, making Bioconductor more powerful for researchers working with complex data. Thanks to funding from CZI EOSS 6, Bioconductor is developing better support for maintainers authoring GPU-capable packages, including adding a new Nvidia GPU build machine maintained at the University of Padova, new release and devel (Nvidia) GPU software builds, GPU-aware containers, and a biocViews GPU term. This post shares these new resources and how Bioconductor package maintainers can take advantage of them.
GPU Support
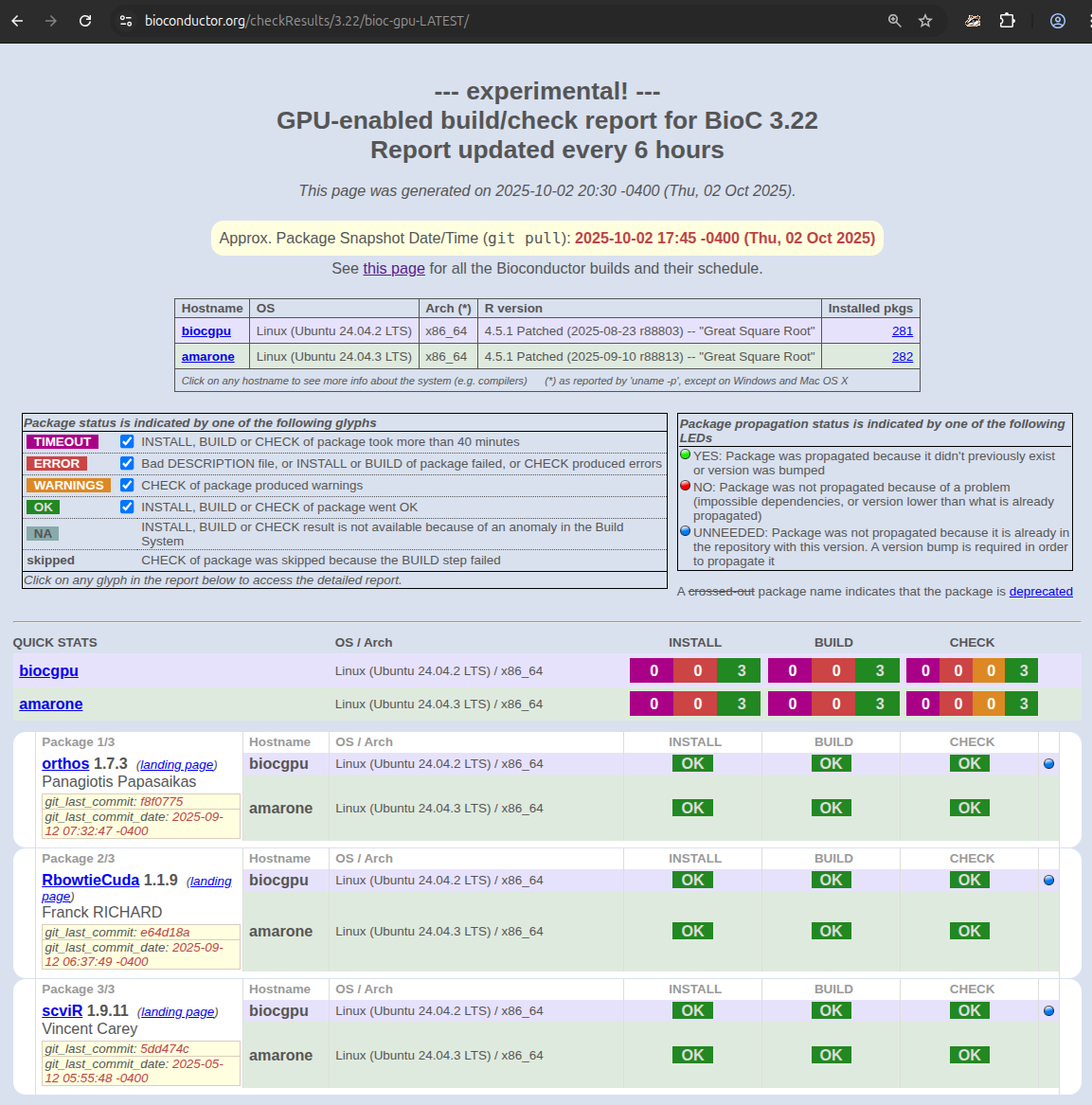
The BBS GPU Software reports are available at
Looking at the reports, you’ll notice three new build nodes:
- biocgpu in Jetstream2
- amarone at the University of Padova
- kakapo1 at the Dana-Farber Cancer Institute
The Nvidia CUDA compiler driver and the Nvidia System Management Interface for each machine appears at the bottom of their node info reports. Both amarone and kakapo1 use a BBS-like container based on Nvidia’s CUDA container available to maintainers.
Additionally, a biocViews term GPU has been added.
How to take advantage of new GPU support
Help users find your GPU-capable package
Opt in to the BBS GPU Software builds
The GPU Software builds, like the Long Tests builds, require a .BBSoptions file at the top level of a package repository with a GPU_reliance setting.
GPU-optional packages
If a package supports GPU if available, maintainers can opt in to the GPU software build by setting GPU_reliance to optional:
GPU_reliance: optionalMake sure you have tests that utilize GPUs if available.
GPU-required packages
Maintainers creating packages requiring GPUs in order for the package to be build should set GPU_reliance to required:
GPU_reliance: requiredSee Advanced Build Options for information.
Future work
More work is being done to understand how we can make the best use of GPUs within Bioconductor by developing a package to understand GPU usage and propose best practices, but we also need a better understanding of what packages are using GPUs and what our community needs. Learn more about the project at https://waldronlab.io/czieoss6-biocgpu/.
© 2025 Bioconductor. Content is published under Creative Commons CC-BY-4.0 License for the text and BSD 3-Clause License for any code. | R-Bloggers